Click-&-Go® Dde Protein Enrichment Kit
* for enrichment of azide-modified proteins *
Product No. 1444
Introduction
Click-&-Go® Dde Protein Enrichment Kit provides all the necessary reagents to perform enrichment of azide-modified proteins through conventional biotin-streptavidin affinity purification. The kit includes a cleavable Dde Biotin Alkyne that allows for release of either captured proteins for intact protein analysis or on-beads digestion followed by the release of peptides for subsequent downstream analysis by mass spectrometry (Figure 1). Captured biomolecules can be released under mild conditions, 2% aqueous hydrazine. Sufficient materials are supplied for 25 enrichments based on the protocol below. The kit provides azide labeled BSA as a positive control.
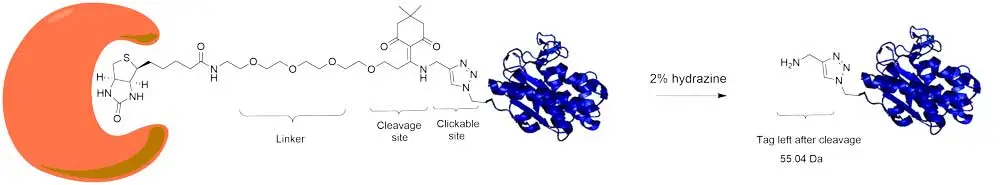
Figure 1. Schematic representation of Dde biotin probe cleavage.
Kit Contents
| Component | Concentration | Amount | Storage | Stability |
|---|---|---|---|---|
| Streptavidin Agarose Resin (Component A) | 50% slurry | 2.5 mL | 4°C | Stable for at least 12 months when stored as directed. |
| DADPS Biotin Alkyne (Component B) | — | 1 vial | 4°C | |
| Copper (II) Sulfate + Protectant (Component C) | 100 mM | 250 μL | 4°C | |
| Reducing Agent (Component D) | — | 100 mg | 4-30°C | |
| Azide Labeled BSA (Component E) | — | 0.5 mg | 4°C | |
| 5% Formic Acid (Component F) | 5% | 3.0 mL | 4-30°C |
Materials Required for Enrichment but Not Provided
- 5-20 mg of cell or tissue extract containing azide-modified biomolecules
- Protease Inhibitors (e.g., Sigma P8340)
- High-speed microcentrifuge
- Unlabeled cells or tissue containing the same relative amount of protein (negative control)
- 1.5 ml microfuge tubes
- 1% SDS in 50 mM Tris-HCl
- Solvents: methanol, DI water
- Probe sonicator or endonuclease such as Benzonase®
Materials Required but Not Provided for On-resin Digestion with Protease.
- DTT, Iodoacetamide
- 8 M Urea/100 mM Tris, pH 8
- Digestion buffer
- Mass spectrometry-grade Trypsin
- C-18 desalting cartridges
- TFA, acetonitrile
- Vacuum concentrator
- Heat block
Additional information
- Concentration of an alkyne biotinylation reagent in click labeling reaction may range from 2 μM to 40 μM. Concentrations below or above this range are also possible, and should be optimized per the specific application. We recommend starting with a 40 μM concentration of alkyne reagent, and titrating this amount down in case of high background or up in case of low reaction efficiency. The kit provides sufficient amount of DADPS Biotin Alkyne reagent to perform 25 labeling reaction using up to 100 μM DADPS Biotin Alkyne concentration.
- Caution: copper (II) sulfate solution is harmful to aquatic organisms and can cause damage to aquatic environments. Avoid release into the environment. Refer to MSDS.
Material Preparation
| High Capacity Streptavidin Agarose Resin (Component A) | Ready to use. Stable for 1 year when stored at 4°C. |
| DADPS Biotin Alkyne (Component B) | Dissolve DADPS Biotin Alkyne reagent in 1.25 mL of DMSO. After use, store unused detection reagent stock at -20°C for up to 1 year. |
| Copper (II) Sulfate (Component C) | Ready to use. Stable for 1 year when stored at 4°C. |
| Reducing Agent (Component D) | Prepare only as much of Reducing Agent (Component D) solution as necessary for that day’s experiment and use on the same day. Weigh 20 mg of Reducing Agent (Component D), add 100 μL of deionized water, vortex until completely dissolved. |
| Positive Control Azide Labeled BSA (Component E) | Reconstitute lyophilized BSA Azide in 500 μL of 0.5x PBS to obtain 1 mg/ml solution. Add 10 μL BSA Azide (Component E) to 1000 μL of unlabeled cell lysate with proteins concentration of 1 mg/mL. This mixture will contain 1% of BSA Azide. The amount of BSA Azide in the positive control is application dependent and can be adjusted to more closely mimic the expected amount of the alkyne labeled proteins in cell lysate. |
1. Cell Lysate Preparation
Do not use DTT, TCEP, or β-mercaptoethanol because they will reduce the azide. Do not use EDTA or any other chelators because they will inhibit click reaction.
- Prepare the lysis buffer by adding protease and phosphatase inhibitors at appropriate concentrations to 1% SDS in 50 mM Tris-HCl, pH 8.0. Alternate lysis protocols (e.g., RIPA buffer, high-salt extraction) are compatible with downstream enrichment.
Note: Protease and phosphatase inhibitors are optional but recommended to ensure sample integrity. - For adherent cells, add 500 μL lysis buffer per 100 mm plate or 200 μL lysis buffer per well of a 6-well plate to the labeled cells. If adding the lysis buffer directly to the plate, tap or rotate the plates so the lysis buffer covers the bottom surface of the plate.
For suspension cell pellet, add 50 μL lysis buffer per 1 × 106 cells. - Incubate the cells for 15–30 minutes on ice, and then tilt the plates and pipet the lysate into a 1.5 mL microcentrifuge tube. If the lysis buffer does not contain Benzonase® endonuclease, the lysate may be very viscous due to the DNA from the lysed cells.
If using Benzonase® endonuclease, proceed to step 1.5. - Sonicate the lysate with a probe sonicator to solubilize the proteins and disperse the DNA.
- Rotate end-over-end the lysate for 5 minutes.
- Centrifuge for 10 minutes at 13,000-20,000 g at 4°C.
- Transfer the supernatant to a clean tube and determine the protein concentration using the BCA Assay or another method. Ideally, the protein concentration should be 1–2 mg/mL.
- The protein sample is now ready for click labeling reaction with DADPS Biotin Alkyne.
2. Biotinylation of Proteins by Click Reaction
This protocol provides guidelines for the click reaction using 1 mL of cell lysate. However, it can be suited for smaller or larger volumes with adjustments of volumes up or down accordingly.
The concentration of DADPS Biotin Alkyne reagent in this protocol is set to 40 μM. The concentrations of azide reagent may range from 2 μM to 80 μM, below or above this range are also possible, and should be optimized for each sample type. For labeling click reaction using higher concentration of DADPS Biotin Alkyne increase volume of DADPS Biotin Alkyne solution added at Step 2.1 leaving volumes of all other reagents unchanged.
- For each protein lysate sample, add the following to a 1.5 mL microfuge tube, then vortex briefly to mix.
• 10 μL DADPS Biotin Alkyne DMSO solution.
• 10 μL Copper (II) Sulfate + Protectant, vortex briefly to mix.
• 10 μL Reducing Agent to initiate click reaction, vortex briefly to mix. - Vortex briefly to mix. This pale blue solution turns colorless after addition of Reducing Agent.
- Add a solution from Step 2.1 to 1000 μL of cell lysate.
- Vortex continuously or rotate end-over-end for 90 minutes at room temperature.
- Add the labeling reaction to 4 mL methanol and 1 mL of Chloroform, vortex briefly. Add 3 mL of water, vortex briefly.
Note: cold (-20°C) acetone (4 mL) can be used in place of methanol:chloroform:water mixture. - Centrifuge for 10 minutes at 13,000-20,000 g at 4°C. Carefully remove upper aqueous layer without disturbing interface layer containing proteins.
- Add 450 μL of cold methanol, vortex briefly.
- Centrifuge for 5 minutes at 13,000-20,000 g to pellet protein. Carefully, remove and discard supernatant.
- Repeat Steps 2.7-2.8.
- Open the lid to microfuge tube and allow protein pellet to air-dry for at least 15 minutes. Do not overdry the pellet!
- Cap and store labeled sample at -20°C until ready for use.
3. Binding Biotinylated Proteins to Streptavidin Agarose Resin
- Resuspend air-dried protein pellets (from Step 2.12) by bath sonication in 800 μL of resuspension buffer (50 mM Tris, 150 mM NaCl and 1% SDS). Other denaturing pull down buffers (for example 6 M urea, 2 M thiourea, and 10 mM HEPES) are also compatible with downstream analysis.
- Mix 50% Streptavidin Agarose slurry (Component A) until the resin is completely resuspended.
- Transfer 100 μL of 50% slurry (50 μL of settled resin) to a microfuge tube. Use a 200 μL pipet with 4-5 mm cut tip to ensure that the resin is transferred properly. Wash streptavidin agarose resin two times with 1 mL of PBS and one time with 1 mL of resuspension buffer.
- Resuspend streptavidin agarose resin in 200 μL of resuspension buffer, and add the slurry to the solubilized proteins.
- Incubate samples on a rotator for 2 h.
- Wash beads four times with resuspension buffer (1 mL) and two times with PBS buffer (1 mL).
Stringency of washing may be increased (e.g., 1% SDS in 8 M urea washes) if high protein background is observed by MS or Western blot. - The resin now contains bound biotin labeled proteins and is ready for cleavage/elution or on-beads digestion.
4. Release of Intact Proteins
- Resuspend the beads from Step 3.6 in 100 μL of 5% formic acid in water.
- Incubate for 30 min at room temperature.
- Centrifuge beads for 5 min at 7000 g, and collect the eluent.
- Optional: Neutralize 5% formic acid by addition of 100 μl of 1 M (or 20 μl of 5 M) solution of sodium hydroxide.
Note: step 4.4 can be replaced by desalting.
5. On Beads Trypsin Digestion of Enriched Proteins
- Resuspend beads from Step 3.6 in 500 μL of 10 mM DTT in PBS, vortex briefly.
- Heat to 70°C on a heat block for 15 minutes, and then cool to room temperature for 15-30 minutes.
- Centrifuge resin for 5 minutes at 2000 g, aspirate the supernatant to waste taking care not to aspirate the resin.
- Add 1 mL 40 mM iodoacetamide solution to the resin, vortex to resuspend the resin, incubate the reaction in the dark for 30 minutes at room temperature.
- Pellet the beads by centrifugation (2000 g, 3 min) and wash once with PBS (1 mL).
- Pellet the beads by centrifugation (2000 g, 3 min).
- Add 300 μL of digestion buffer (100 mM Tris, 2 mM CaCl2, 0.5 M urea) to the resin.
- Add 3 μg trypsin from a stock solution to the resin slurry, gently mix the slurry, then incubate at 37°C for 6-12 hours with over-the-end rotation.
Note: Alternative proteases and trypsin digestion procedures are also compatible with this protocol and may be used to increase peptide coverage. - Pellet the beads by centrifugation (2000 g, 3 min), and collect the supernatant digest.
- Wash the beads with PBS (200 μL) and water (2 × 200 μL). Keep the resin.
- Combine the washes with the supernatant digest to form the “trypsin fraction”.
- Concentrate the “trypsin fraction” to dryness using a speedvac set to 40°C.
- The “trypsin fraction” may be stored for at least 12 months at –80°C.
6. Cleavage and Recovery of Peptides
- Resuspend the beads from Step 5.10 in 100 μL of 5% formic acid in water.
- Incubate for 30 min at room temperature.
- Centrifuge beads for 2 min at 2000 g, and collect the eluent.
- Resuspend the beads in 200 μL of 50% acetonitrile–water, centrifuge beads for 2 minutes, and collect the eluent.
- Combine all eluents.
- Concentrate the “cleavage fraction” to dryness using a Speedvac heated to 40°C. Store at –20°C until desalting and MS analysis.
- For long term storage, the “cleavage fraction” may be stored for at least 12 months at –80°C.
7. Preparation of Digest for Mass Spectrometry Analysis
The following protocol is provided for desalting digested peptides (from Step 5.13) using C-18 desalting cartridges (e.g. Waters WAT036820). Sample recovery for typical peptides is > 85%, but could be as low as 35% for hydrophilic peptides. Other desalting protocols for sample preparations for MS also can be used.
For desalting peptides released from the beads after Step 6.8 we recommend using pipette-tip columns such as ZipTip C18 P10 or Pierce C18 Tips using manufacture recommended protocol.
- Acidify the diluted digest by adding 10 μL of TFA.
- Desalt the digest on a C-18 cartridge using vacuum or gravity flow, allowing each solution to completely flow through the cartridge before adding the next solution.
a. Add 1 mL of 50% acetonitrile/0.1% TFA to the cartridge and discard the effluent.
b. Add 1 mL of 0.1% TFA to the cartridge and discard the effluent. Repeat one more time.
c. Add the acidified, diluted digest to the cartridge and discard the effluent.
d. Add 1 mL of 0.1% TFA to the cartridge and discard the effluent. Repeat one more time.
e. Place a clean 1.5 mL tube below the C-18 cartridge outlet.
f. Elute the peptides into a clean 1.5 mL tube by adding 700 μL of 50% acetonitrile/0.1% TFA to the C-18 cartridge. - Dry the eluate containing the desalted peptide digest in a vacuum concentrator. Store at -20°C until ready for MS analysis.
Troubleshooting
| Problem | Possible Cause | Solution |
|---|---|---|
| Low yield of enriched peptides | Inefficient protein click labeling with biotin or low abundance of azide-tagged proteins | Increase lysate concentration (use more cells) or pre-enrich the proteins (e.g. soluble lysate, membrane lysate, lectin enrichment, etc.). Increase concentration of biotin labeling reagent |
| Residual biotin reagent remained in cell lysates after click labeling | Perform two additional washes with methanol | |
| Inefficient digestion of resin-bound proteins | Use high quality trypsin | |
| High background with unlabeled control cells | Insufficient washing of resin | Increase column washes Use only high purity reagents Prepare filtered buffers fresh Ensure proper preparation of copper catalyst solution |
| High no-specific click labeling reaction | Decrease concentration of biotin labeling reagent |

